How to make an Atlas
This document will describe the steps used to create the most recent version of the DK atlas.
The data used to create this atlas was initially made by several anatomists working at the DK lab at UCSD. Structures were drawn onto full resolution mouse brain images and the polygon vertices were then stored as CSV files on the network filesystem. Three different mouse brains were used and they are collectively known as the foundation brains and they are listed below:
MD585
MD589
MD594
MD589 is used as the reference brain. There are 51 brain structures that were drawn by the anatomists. Not every anatomist drew every structure on each of the three brains. This results in a varying number of drawn polygons per structure per brain. Each brain has it’s own CSV file and the process parses each CSV file. The first 10 rows of MD589 are show below:
name creator orientation section side time_created vertices
VCA yuncong sagittal 367 R 9152016033619 [[15450.64315379 13041.15143737]\n [15292.3766...
IO yuncong sagittal 260 R 9122016211223 [[25199.46145603 17916.30221869]\n [24976.1090...
LRt yuncong sagittal 286 R 9152016024654 [[29069.49187508 17203.67921491]\n [28909.8937...
LRt yuncong sagittal 276 R 9152016024654 [[29219.12776439 16422.80664145]\n [29084.8059...
SC yuncong sagittal 202 S 5312018021355 [[21584.44352233 7847.78193722]\n [21497.5691...
Looking at the data above, we can see that for each section of each structure, multiple anatomists may have drawn polygons. These polygons are averaged together on each section and then these sections are concatenated to form a volume. For each structure, the minimum x,y, and z values are found. These values are used to create a bounding box with the upper left corner of the box being the origin. The origin is saved and the mean of all the origins is subtracted from each origin. This is where the bounding box is placed within the atlas. For each brain section, the set of points is drawn on this bounding box. All the sections are then appended to form the box and within the box is the structure. The box is saved as a numpy array with either zeroes or the value 255. Centers of mass are calculated for each of the drawn volumes and these are then used to create a rigid transformation matrix that is used to align the drawn polygons to the reference brain. This process now yields the origin of the bounding box, the actual 3D volume and then the center of mass. All this data is initially stored on the filesystem and the centers of mass are also stored in the database.
In the following example, the SC structure has 3 drawn volumes that now need to be merged. They are first aligned to the largest of the 3 volumes using a rigid transformation.
Here is a preview of what the structure superior colliculus (SC) looks like at the mid z value:
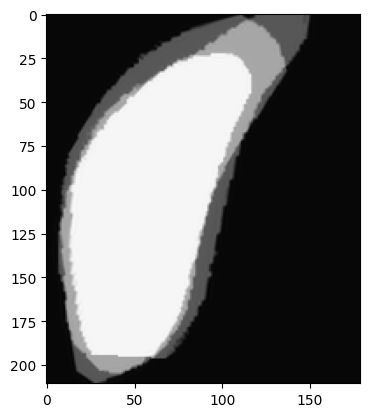
Now that they are aligned, the following steps are peformed to get a single smooth volume:
The three volumes are smoothed together with a gaussian filter with a large standard deviation.
This results in a volume of floats. An abitrary value of 150 is used where the values above 150 are set to the Allen ID and all other values are set to zero.
The 3D volume is now set to be a binary volume with values of 0 and the Allen ID. Since the Allen IDs are large integers, the volume is saved as a 32bit Integer.
The structure superior colliculs (SC) looks like this after using smoothing and merging:
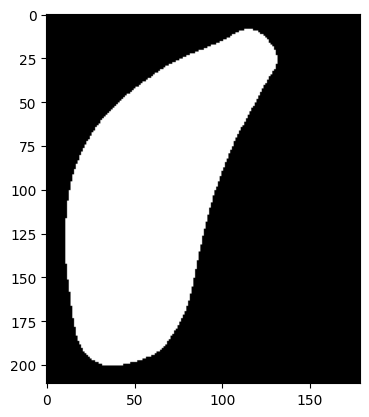
The same process is repeated for the remaining 50 structures.
Structures not in the foundation brains can also be merged into the atlas. These are created from polygons drawn in neuroglancer by the anatomists. The polygons are stored in the database, retreived and the same process as the foundation brain structures is repeated. One problem is getting a rigid transformation between the neurotrace brains and the foundation brains. An Elastix rigid transformation is calculated using the full 3D volume of a 10um isotropic MD589 brain and the neurotrace brain. This transformation is then applied to the neurotrace data points which puts them into the same coordinate system as MD589.
We now have all the merged brain structures in MD589 10um isotropic space. The next step is to register this atlas with the Allen reference atlas. We use the Allen SDK (https://allensdk.readthedocs.io/en/latest/) to get the centers of mass of common structures between the DK atlas and the Allen reference atlas. One problem with this approach is structures have different names in the Allen SDK and the DK atlas. Another problem is the Allen atlas breaks some structures into sub structures (e.g. the SC structure). Another issue is the DK atlas differentiates between left and right structures while the Allen does not.
A rigid transformation is calculated between the DK atlas and the Allen reference atlas using the centers of mass of the common structures. Results are show below and all values are in micrometers:
structure |
AtlasV8 |
Allen |
transformed |
difference |
sumsquares |
|---|---|---|---|---|---|
IC |
[9901. 3173. 6876.] |
[10400. 2325. 5675.] |
[10413. 2329. 5665.] |
[ 13. 4. -10.] |
17.1672 |
5N_R |
[9728. 5471. 8282.] |
[10193. 5286. 7293.] |
[10163. 5314. 7266.] |
[-30. 28. -27.] |
49.3379 |
PBG_R |
[8927. 4396. 8771.] |
[9401. 3846. 7833.] |
[9442. 3893. 7840.] |
[42. 47. 7.] |
63.1534 |
LRt_L |
[12146. 6651. 5841.] |
[12344. 6991. 4393.] |
[12418. 6924. 4437.] |
[ 73. -67. 44.] |
108.5987 |
3N_L |
[8688. 4514. 6769.] |
[9102. 3794. 5521.] |
[9180. 3874. 5529.] |
[77. 80. 8.] |
111.3796 |
6N_L |
[10292. 5495. 6547.] |
[10771. 5215. 5291.] |
[10685. 5290. 5264.] |
[-86. 76. -27.] |
117.9381 |
LRt_R |
[12093. 6497. 8060.] |
[12344. 6991. 6993.] |
[12402. 6877. 7000.] |
[ 58. -113. 7.] |
127.8019 |
SNC_R |
[7827. 5408. 8082.] |
[8356. 5123. 7092.] |
[8326. 4994. 7037.] |
[ -30. -129. -56.] |
143.528 |
3N_R |
[8664. 4544. 7053.] |
[9103. 3794. 5864.] |
[9159. 3928. 5857.] |
[ 56. 134. -8.] |
145.6706 |
Sp5I_L |
[11888. 5951. 5153.] |
[12107. 5872. 3748.] |
[12190. 5960. 3650.] |
[ 83. 88. -98.] |
155.4577 |
5N_L |
[9714. 5496. 5565.] |
[10193. 5287. 4092.] |
[10113. 5154. 4130.] |
[ -80. -133. 38.] |
159.6781 |
4N_L |
[9053. 4525. 6658.] |
[9589. 3778. 5432.] |
[9531. 3924. 5401.] |
[-58. 146. -32.] |
160.4182 |
6N_R |
[10276. 5504. 7272.] |
[10771. 5215. 6094.] |
[10678. 5350. 6100.] |
[-93. 136. 6.] |
164.886 |
7N_L |
[10494. 6581. 5850.] |
[10853. 6775. 4340.] |
[10824. 6640. 4449.] |
[ -29. -136. 109.] |
176.2443 |
SNR_R |
[8114. 5462. 8325.] |
[8440. 5167. 7353.] |
[8604. 5113. 7316.] |
[164. -54. -38.] |
176.4407 |
Sp5I_R |
[11922. 5660. 8602.] |
[12107. 5872. 7638.] |
[12281. 5836. 7633.] |
[174. -36. -5.] |
178.0143 |
4N_R |
[9015. 4522. 7130.] |
[9587. 3775. 5953.] |
[9501. 3948. 5945.] |
[-86. 173. -8.] |
193.4545 |
SC |
[8584. 3186. 6937.] |
[9140. 2388. 5692.] |
[9139. 2193. 5735.] |
[ -1. -195. 42.] |
199.4932 |
SNC_L |
[7926. 5467. 5747.] |
[8354. 5125. 4294.] |
[8388. 4918. 4340.] |
[ 34. -207. 46.] |
214.9368 |
DC_L |
[10850. 5166. 4883.] |
[11270. 5002. 3229.] |
[11217. 4824. 3346.] |
[ -53. -178. 117.] |
219.2414 |
Amb_R |
[11344. 6274. 8158.] |
[11881. 6618. 7062.] |
[11689. 6512. 7116.] |
[-192. -105. 54.] |
225.6861 |
7N_R |
[10551. 6380. 8110.] |
[10853. 6775. 7045.] |
[10917. 6550. 7059.] |
[ 64. -225. 14.] |
234.7462 |
Amb_L |
[11360. 6440. 5677.] |
[11883. 6618. 4324.] |
[11665. 6552. 4250.] |
[-219. -65. -74.] |
239.7618 |
Sp5O_L |
[11039. 5937. 5174.] |
[11139. 5912. 3787.] |
[11370. 5842. 3674.] |
[ 231. -70. -112.] |
266.5864 |
LC_L |
[10106. 4905. 6053.] |
[10710. 4278. 4732.] |
[10524. 4488. 4699.] |
[-185. 210. -33.] |
281.6734 |
PBG_L |
[9074. 4354. 5046.] |
[9403. 3844. 3551.] |
[9538. 3597. 3542.] |
[ 135. -247. -10.] |
281.8039 |
VLL_L |
[9017. 5800. 5423.] |
[9464. 5176. 3890.] |
[9424. 5446. 3964.] |
[-40. 270. 74.] |
283.0251 |
Sp5C_L |
[12605. 6033. 5519.] |
[12833. 5896. 3980.] |
[12884. 6174. 4071.] |
[ 52. 278. 91.] |
297.3531 |
7n_R |
[10161. 5724. 7969.] |
[10688. 5482. 6677.] |
[10567. 5663. 6902.] |
[-121. 181. 225.] |
313.1624 |
Sp5O_R |
[11062. 5739. 8539.] |
[11138. 5913. 7598.] |
[11445. 5829. 7559.] |
[306. -84. -38.] |
319.9945 |
DC_R |
[10857. 4767. 9084.] |
[11271. 5002. 8156.] |
[11297. 4613. 8197.] |
[ 26. -389. 41.] |
391.6241 |
LC_R |
[9976. 4870. 7679.] |
[10710. 4279. 6652.] |
[10421. 4541. 6576.] |
[-289. 263. -76.] |
397.7449 |
VLL_R |
[8894. 5741. 8414.] |
[9465. 5174. 7495.] |
[9346. 5565. 7416.] |
[-119. 391. -80.] |
416.4021 |
7n_L |
[10155. 5868. 5785.] |
[10687. 5483. 4706.] |
[10526. 5693. 4380.] |
[-161. 210. -326.] |
419.9833 |
SNR_L |
[8274. 5418. 5595.] |
[8441. 5167. 4032.] |
[8724. 4886. 4165.] |
[ 284. -281. 133.] |
420.8795 |
AP |
[12074. 5465. 6772.] |
[12653. 5010. 5692.] |
[12412. 5480. 5523.] |
[-241. 470. -169.] |
554.4442 |
RtTg |
[9011. 6161. 6995.] |
[9503. 5322. 5692.] |
[9423. 6012. 5775.] |
[-80. 690. 82.] |
699.4045 |
With the rigid transformation, we can now create the 10um isotropic atlas. The 10um Allen atlas is of the following size:
x_length = 1320
y_length = 800
z_length = 1140
We extend the x and y axes to accomodate the brain stem:
x_length = 1820
y_length = 1000
z_length = 1140
The origin of each structure is retrieved and the new bounding box is calculated from the sizes listed above. This origin is then transformed by the rigid transformation matrix between the DK atlas and the Allen reference atlas. Each structure is then placed within the super volume and that super volume is then used to create a Neuroglancer dataset.
The atlas can be view in Neuroglancer at the following URL: